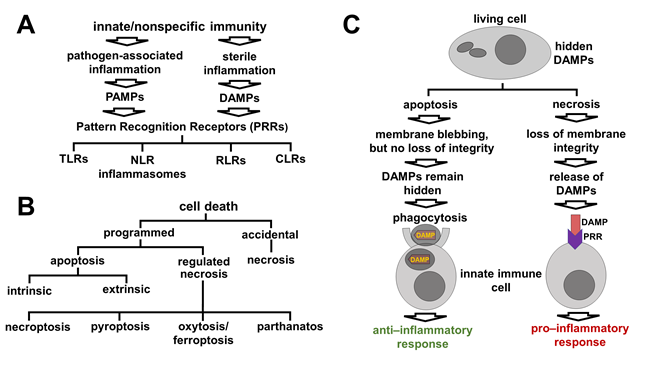
Though much progress has been made in the understanding of signaling cascades driving retinal development and pathology, the role of epigenetic mechanisms remains to be explored as meticulously. To fill this gap, we evaluated the contribution of the DNA methylation and DNA demethylation pathways to retinal development and pathology in recent studies (Fig. 3). We discovered that the promoters (a region of DNA where the transcription of a gene is initiated) of many mutated in retinitis pigmentosa (RP) and related photoreceptor dystrophies genes were highly methylated (hypermethylated) in DNA isolated from fetal retinas and retinal progenitor cells (RPCs; e.g., USH2A, RHO, PRPH2, EYS, AIPL1, CNGB1, IMPG1, IMPG2, NR2E3, PDE6A, PDE6G, PDE6C, PDE6H, RBP3, RP1). The methylation of these promoters was significantly reduced during RPC differentiation into photoreceptors and accompanied by an increased expression of the corresponding genes. It is generally accepted that DNA methylation in promoter regions silences gene expression, while DNA demethylation should occur to allow gene expression. Hence, unsuccessful demethylation of the promoters of the genes listed above during RPC differentiation into photoreceptors may reduce or even eliminate their activity, leading to photoreceptor dystrophies without any mutations in the genomic DNA (Fig. 4). Thus, not only mutations in DNA but also retina-specific epigenetic changes in the DNA may contribute to the pathogenesis of RP and related diseases, indicating the importance of understanding the DNA demethylation pathway during photoreceptor development. The ten–eleven translocation (TET) protein family has a vital role in DNA demethylation and regulates eye development and neurogenesis in various species. Our data and the results of other laboratories indicate that the TET-dependent DNA demethylation pathway controls photoreceptor development. The objectives of this project are to gain a detailed understanding of how the TET-driven DNA demethylation pathway specifies the differentiation of RPCs into photoreceptors, and to investigate how irregularities in its activity lead to photoreceptor death and retinal degeneration.
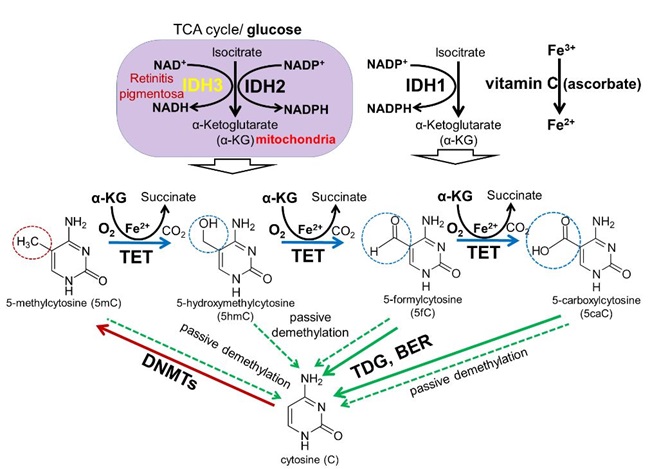
Figure 3. DNA methylation and demethylation pathways: During development, patterns of methylated cytosines are established by the de novo methyltransferases Dnmt3a/Dnmt3b, and subsequently preserved through cell divisions by Dnmt1. The TET family promotes DNA demethylation by oxidizing 5mC to produce 5hmC, 5fC, and 5caC in DNA. Oxidized derivatives of 5mC inhibit Dnmt1, promoting passive DNA demethylation (dashed lines). 5fC and 5caC are directly excised by thymine DNA glycosylase (TDG) to generate abasic sites triggering base excision repair (BER) pathway activation followed by replacement of the abasic sites with unmodified cytosines.
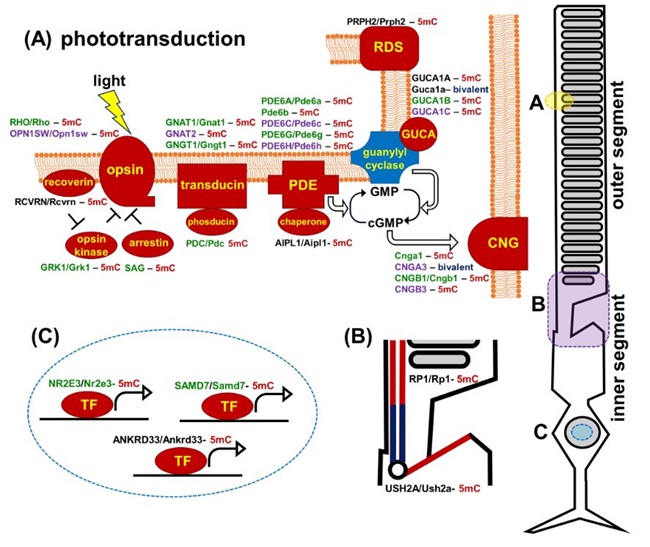
Figure 4. Promoters of human and mouse genes coding molecular components of the photoreceptor outer segment (phototransduction cascade) and inner segment are mostly hypermethylated in embryonic retinas/RPCs (the corresponding proteins are marked in red). The genes are colored green for rods, purple for cones, and black indicates expression in both rods and cones. (5mC- 5-methylcytosine, gene with hypermethylated promoter; bivalent- bivalent promoter)